The geNomad framework for classification and annotation
geNomad employs a hybrid strategy to plasmid and virus identification that mixes an alignment-free classifier (sequence department) and a gene-based classifier (marker department) to enhance classification efficiency by capitalizing on the strengths of every classifier. geNomad’s framework consists of 5 levels (Fig. 1a): (1) alignment-free classification within the sequence department; (2) sequence annotation and gene-based classification within the marker department; (3) aggregation of the department scores; (4) rating calibration; and (5) output technology.
a, geNomad processes user-provided nucleotide sequences by way of two branches. Within the sequence department, the inputs are one-hot encoded fed to an IGLOO neural community, which scores inputs primarily based on the detection of non-local sequence motifs (A1 I). Within the marker department, proteins encoded by the enter sequences are annotated utilizing markers which can be particular to chromosomes, plasmids or viruses (A1 II). A set of numerical options is then extracted from the annotated proteins and fed to a tree ensemble mannequin, which scores the inputs primarily based on their marker content material. Subsequent, the scores supplied by each branches are aggregated by weighing the contribution of every department primarily based on the frequency of markers within the sequence (A2). Aggregated scores can then be calibrated to approximate possibilities in a course of that leverages the pattern composition inferred from the classification of sequences from the identical batch (A3). Lastly, classification outcomes are summarized and offered along with further information, akin to virus taxonomy, gene operate and the inferred genetic code (A4). b, The sequence department relies on the IGLOO structure, which makes use of convolutions to provide a characteristic map from a one-hot encoded enter. Patches encoding non-local relationships inside the sequence are then generated by slicing the characteristic map. Lastly, these patches are used as an consideration matrix to provide a sequence illustration from the characteristic map. c, The relative contribution of the marker department (y axis, quantified utilizing SHAP) will increase because the marker frequency (fraction of genes assigned to a marker) within the sequence will increase. d, Calibration curves of pre-calibration (left) and post-calibration (proper) scores, displaying that pattern composition can be utilized to map classification scores to precise possibilities. The x axis represents scores averaged throughout a number of bins; the y axis represents the fraction of positives in every bin; the 45° dashed line represents an ideal calibration situation. freq., frequency; MAE, imply absolute error of the scores relative to the true possibilities.
To determine sequences of plasmids and viruses in an alignment-free method, geNomad’s sequence department makes use of a neural community mannequin that may classify the sequences from their nucleotide make-up alone (Fig. 1a, field A1 I). To course of enter sequences, geNomad employs an encoder primarily based on the IGLOO structure10, which is ready to extract patterns which can be helpful for classification from the nucleotide sequences and encode them into an embedding area (Fig. 1b and Prolonged Knowledge Fig. 1). This structure has demonstrated superior efficiency in comparison with conventional options (akin to recurrent and convolutional neural networks) when utilized to sequence information, because it gathers info from non-local relationships throughout the sequence to create a world illustration10,11.
To categorise sequences primarily based on their gene content material, geNomad’s marker department predicts and annotates the proteins encoded by enter sequences utilizing a set of customized markers (Fig. 1a, field A1 II). To foretell proteins, geNomad makes use of a modified model of the Prodigal12 software program known as prodigal-gv, which we developed to permit computerized detection of recoded TAG cease codons (widespread in Crassvirales phages13) and annotation of TATATA motifs which can be regularly discovered upstream of coding sequences of Nucleocytoviricota viruses14. Predicted proteins are then queried in opposition to a set of 227,897 protein profiles—particular to chromosomes, plasmids or viruses (Fig. 2)—utilizing MMseqs2 (ref. 15) protein profile search. Subsequent, geNomad computes a complete of 25 numeric genomic options that summarize the sequence construction (for instance, gene density and strand swap fee), RBS motifs (for instance, TATATA motif frequency) and marker content material (for instance, frequency of chromosome, plasmid and virus markers) of the enter sequences (Supplementary Observe 1 and Supplementary Desk 1). These options are then fed to a tree ensemble classification mannequin, which outputs the boldness scores for every class.
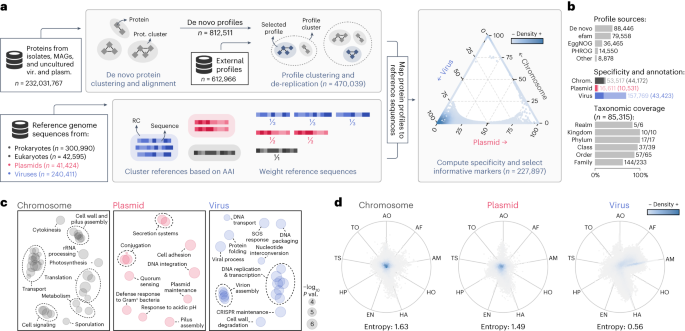
a, Protein sequences from genomes and metagenomes have been clustered and aligned to provide de novo protein profiles. De novo profiles and profiles obtained from public databases have been then clustered, and cluster representatives have been chosen to cut back redundancy. In parallel, reference chromosome, plasmid and virus sequences have been clustered into RCs. Sequences have been then weighed in such a means that the sum of the weights inside every RC was fixed. Consultant protein profiles have been mapped to reference sequences, and chromosome-, plasmid- and virus-specificity metrics have been computed for every profile primarily based on the weighed variety of hits to sequences of every class. Markers that have been extremely particular to one of many three courses have been then chosen. The place of every chosen marker (circles) within the ternary plot is decided by its specificity, and the colours characterize the marker density in a area. b, Bar plots displaying: the sources of the chosen profiles (higher plot); the entire variety of markers (mild shades) and the variety of functionally annotated markers (darkish shades) for every class (center plot); and the fraction of ICTV taxa lined by the taxonomically informative markers at every rank. c, Multidimensional scaling of semantic similarities of the GO phrases enriched in chromosome (left), plasmid (middle) and virus (proper) markers. Labels of associated phrases have been aggregated for readability. Semantic similarities have been computed with REVIGO. d, RadViz visualizations of the relative frequencies of geNomad markers throughout distinct ecosystems. Every marker is represented by a circle, and the colours depict the marker density inside a area. The place of the markers within the plot is decided by their frequency in every atmosphere. Markers near the middle of the plot have been present in related frequencies throughout all ecosystems. Median entropies of the ecosystem distributions are proven beneath the plots. AF, aquatic (freshwater); AM, aquatic (marine); AO, aquatic (different); EN, engineered; HA, host-associated (animals); HO, host-associated (different); HP, host-associated (crops); TO, terrestrial (different); TS, terrestrial (soil).
From the outputs produced by the sequence and marker branches, geNomad generates an aggregated classification that leverages the strengths of every strategy. That is achieved by way of an consideration mechanism that consists of a linear mannequin that weighs the branches primarily based on the frequency of chromosome, plasmid and virus markers within the enter sequence (Fig. 1a, field A2). The eye mechanism works in such a means that the contribution of the marker department goes larger because the fraction of genes which can be assigned to markers will increase (Fig. 1c). This permits geNomad to make the most of each marker-based and alignment-free classification approaches in a principled method.
Throughout inference, a classification mannequin assigns a rating to every prediction, indicating the diploma of confidence in that prediction, with larger values representing extra assured predictions. Nonetheless, these scores don’t replicate the true possibilities of the predictions being right, as classification fashions will exhibit various false discovery charges (FDRs) when classifying samples with distinct underlying composition (Supplementary Observe 2 and Prolonged Knowledge Fig. 2). To handle this, we devised an elective calibration mechanism in geNomad that leverages pattern composition information to approximate the true underlying possibilities. (Fig. 1a, field A3, and Fig. 1d). The calibrated scores produced by geNomad supply customers two advantages: (1) estimated possibilities can be utilized to compute FDRs, permitting customers to make extra knowledgeable selections (for instance, setting a threshold to realize a desired proportion of false positives); and (2) improved classification efficiency by adjusting the assigned labels of some sequences after calibrating scores (for extra particulars, see ‘geNomad precisely identifies plasmids and viruses’ part).
Sequences categorized as viral with geNomad’s markers are then assigned to taxa outlined by the Worldwide Committee on Taxonomy of Viruses (ICTV)16. This course of is made potential by the truth that greater than 85,000 of the markers are particular to a virus taxon (for extra particulars, see ‘A dataset of marker protein profiles’ subsection). In short, geNomad assigns a taxon to every gene annotated with a taxonomically knowledgeable marker. Subsequently, it aggregates the taxonomies of all of the genes inside every scaffold and generates a single consensus lineage for that sequence (Prolonged Knowledge Fig. 3).
Upon completion of its execution, geNomad produces a listing of sequences which were categorized as both plasmids or viruses. This listing could be refined utilizing further user-adjustable filters, such at least rating, most FDR (if rating calibration was carried out), minimal variety of plasmid or virus hallmark genes and most quantity common single-copy genes. The generated output consists of wealthy metadata that may be helpful for downstream evaluation (Fig. 1a, field A4) and the nucleotide and amino acid sequences of the recognized plasmids and viruses.
A dataset of marker protein profiles
geNomad makes use of a marker set of 227,897 protein profiles particular to chromosomes, plasmids or viruses to carry out classification primarily based on gene content material and to offer purposeful info for processed sequences (Fig. 2a). To construct this marker dataset, which covers sequences from uncultured microorganisms and viruses from various environments, we clustered roughly 232 million protein sequences from various sources (see ‘Database of genomic sequences for coaching and benchmarking’ part). The ensuing clusters have been independently aligned, producing 812,511 de novo protein profiles, which have been additional supplemented with 612,966 exterior profiles. To enhance geNomad’s computational effectivity and guarantee broad protection of the gene area, we recognized and eliminated redundant profiles, leading to a group of 470,039 non-redundant profiles (Prolonged Knowledge Fig. 4a,b).
To pick out profiles which can be informative for classification, we computed the specificity of every profile to every one of many focused courses (chromosomes, plasmids and viruses) by mapping them to proteins encoded by reference genomes of each isolate and uncultivated species (Prolonged Knowledge Fig. 4c) and counting the hits to every class. To mitigate the bias ensuing from uneven taxonomic illustration of plasmid and virus sequences in public databases, which favor components infecting a restricted vary of microbes, we downweighted sequences belonging to overrepresented taxa by clustering them into reference clusters (RCs) that group related genomes. We assigned weights to the references in order that the sum of the weights in all RCs was fixed, successfully downweighting sequences inside giant RCs7. After computing specificity, we discarded profiles that have been poorly particular or that matched few proteins, leading to a ultimate set of 227,897 profiles. Many of the markers originated from the de novo protein clustering (38.8%), efam17 (34.9%) and EggNOG18 (16.0%) (Fig. 2b, high, and Supplementary Desk 2). Virus-specific markers dominate the dataset (69.2%), adopted by chromosome-specific markers (23.5%) and plasmid-specific markers (7.3%) (Fig. 2b, center, lighter shades).
geNomad additionally gives detailed taxonomic and purposeful info for organic interpretation of outcomes, enabling thorough evaluation of recognized MGEs. To permit this, markers have been functionally annotated through alignment to the Pfam-A19, TIGRFAM20, KEGG Orthology21 and COG22 databases. In complete, 98,127 (43.1%) markers have been annotated, though the proportion of annotated markers various among the many completely different specificity courses, with chromosome-specific markers having the best annotation fee (82.5%), adopted by plasmid-specific markers (63.4%) and virus-specific markers (27.5%) (Fig. 2b, center, darker shades, and Supplementary Desk 2). Practical enrichment evaluation of the annotated markers (Fig. 2c) revealed that chromosome markers have been related to translation, transport and metabolism capabilities; plasmid markers have been enriched in quorum sensing and motility capabilities; and virus markers have been associated to virus replication and meeting capabilities. A complete of 978 plasmid and 14,635 virus markers have been manually chosen as hallmark markers, as they have been annotated with capabilities associated to core processes, akin to conjugation genes for plasmids and capsid proteins for viruses. To supply further context for MGE analysis, markers have been additionally annotated utilizing databases for particular domains of curiosity (Supplementary Desk 2), ensuing within the identification of 484 markers for genes concerned in conjugation and 382 markers for antimicrobial resistance, annotated by way of alignment with the CONJscan23 and NCBIfam-AMRFinder24 databases, respectively. Lastly, 741 markers for common single-copy genes, that are hardly ever current in MGEs and can assist scale back false positives, have been recognized by way of comparability with profiles from the BUSCO dataset25.
To permit taxonomic task of viruses utilizing geNomad’s markers, virus taxa from the ICTV (Virus Metadata Useful resource model 19) have been assigned to 85,315 markers. The taxonomically knowledgeable markers can be utilized to assign virus sequences to a considerable fraction of the viral taxa as much as the household rank (Fig. 2b, backside), as at the least one marker was assigned to 83.3% of the realms (the one realm lacking is Ribozyviria), 100% of the kingdoms and phyla, 94.9% of the courses, 87.7% of the orders and 61.8% of the households. Most of those markers have been assigned to the Caudoviricetes class (93.1%), which dominates metagenomic information9, however different main taxa, akin to Riboviria (2.8%), Nucleocytoviricota (2.2%) and Monodnaviria (0.7%), are additionally largely lined (Supplementary Desk 2).
Our marker choice course of was designed to maximise the vary of lined uncultivated genomes discovered globally. To evaluate the environmental breadth of geNomad’s markers, we used them to scan a complete of two.3 billion proteins from 28,865 metagenomes and seven,258 metatranscriptomes of varied ecosystems. The ecosystem distributions of the marker courses (chromosome-, plasmid- and virus-specific) have been then evaluated (Supplementary Strategies), revealing that chromosome-specific and plasmid-specific markers are usually not particular to any ecosystem (excessive common entropy of frequencies), whereas virus-specific markers are typically restricted to particular ecosystems (low common entropy of frequencies) (Fig. second). This implies that the gene repertoire of uncultivated viruses is very variable and highlights the significance of incorporating environmental information to cowl a big fraction of the virosphere.
geNomad precisely identifies plasmids and viruses
To judge the classification efficiency of geNomad and evaluate it to different virus and plasmid identification instruments that use completely different approaches for sequence classification (Desk 1), we used take a look at datasets consisting of various sequence fragments with various lengths (Prolonged Knowledge Fig. 5a). To reduce overestimation of geNomad’s efficiency as a result of presence of comparable sequences within the practice and take a look at information, we randomly assigned RCs to 5 completely different information splits and carried out cross-validation utilizing the leave-one-group-out technique (see Strategies for particulars), which pressured sequences from the identical RC to stay collectively in both the practice or take a look at units. Efficiency metrics for all instruments have been measured 5 instances, utilizing every RC because the take a look at set at a time. Further benchmark outcomes are described in Supplementary Observe 3.
By evaluating the classification, measured utilizing the Matthews correlation coefficient (MCC), as a operate of the similarity to the practice information, we discovered that geNomad performs nicely on unseen genomes, regardless that efficiency dropped for sequences that have been extra divergent from the practice information (Prolonged Knowledge Fig. 5b). Evaluation of geNomad’s efficiency on sequences with various marker protection (that’s, fraction of proteins assigned to markers) revealed that even those who have been focused by no or few markers have been nonetheless detected as a result of sequence department of the algorithm (Prolonged Knowledge Fig. 5c). When in comparison with different instruments, geNomad offered superior general classification efficiency throughout all sequence size ranges in each plasmid and virus classification duties (Fig. 3a,b and Supplementary Tables 3 and 4). Such distinction was significantly obvious for brief sequences (<6 kilobases (kb)), the place different instruments confirmed lowered efficiency because of restricted genetic info, whereas geNomad leveraged its intensive marker dataset and alignment-free classification mannequin, guaranteeing excessive sensitivity and precision. This highlights the usefulness of geNomad in metagenomic and metatranscriptomic assemblies, the place most scaffolds are quick.
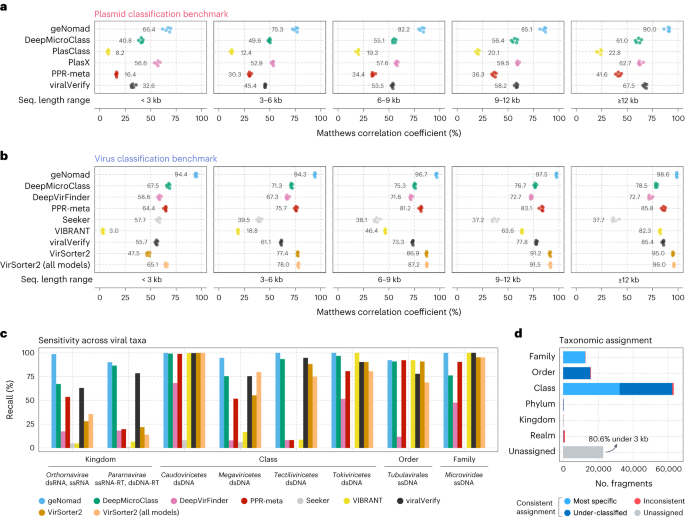
a,b, Classification efficiency of a number of plasmid (a) and virus (b) identification instruments throughout sequence fragments of various size. Efficiency was measured utilizing the MCC. For every sequence vary interval, instruments have been evaluated with 5 completely different take a look at units, every containing the sequences of 1 RC. Coloured circles characterize the performances measured in every take a look at set. Imply values are proven subsequent to the circles. c, Sensitivity of virus identification instruments throughout main viral taxa at completely different ranks. The rating cutoff of every device was decided in order that the FDR was roughly 5%. d, Virus taxonomic task efficiency. Bar lengths characterize the variety of sequence fragments assigned at a given taxonomic rank. Mild blue represents sequences that have been accurately assigned to their most particular rank (as much as the household stage); darkish blue represents fragments that have been assigned to the right lineage however to a rank that’s above its most particular rank; pink represents sequences that have been assigned to the mistaken lineage; and the grey bar represents sequences that have been assigned to any taxon.
geNomad’s calibration mechanism enhances the classification course of by incorporating pattern composition information and assigning estimated possibilities to every sequence, which replicate the probability of the sequence belonging to every class. Our evaluation confirmed that the plasmid classification efficiency elevated with using calibrated scores, significantly for shorter sequences (common ΔMCC: +11.8% for sequences <3 kb; +5.6% for 3–6 kb; and +3.2% for six–9 kb) (Prolonged Knowledge Fig. 5d). We additionally discovered that quick virus sequences benefited from calibration, though the advance was not as pronounced. These outcomes showcase the effectiveness of the launched calibration mechanism for bettering classification high quality.
Plasmid classification is a difficult job as a result of variable genetic make-up of those components, their similarity to different cellular components that may combine into host chromosomes and the dearth of a regular for reporting plasmids in sequencing information. In consequence, most evaluated instruments (DeepMicroClass26, PPR-Meta27, PlasClass28 and viralVerify29) had low common classification precision (11.0–40.1%; Supplementary Desk 3), even when classifying lengthy sequences (Supplementary Desk 4), as they typically produced a excessive variety of false positives that may impression downstream evaluation. In distinction, PlasX7 had excessive precision (81.6%) however low sensitivity (40.5%), which impairs the detection of plasmids in sequencing information. geNomad had the perfect general efficiency by a considerable margin (Fig. 3a; MCC and F1-score in Supplementary Tables 3 and 4), with the best sensitivity (89.8%) and the second highest precision (70.8%), after PlasX. It’s price noting that geNomad’s marker department, which could be run independently, achieved a significantly larger precision than PlasX (91.2%). Analysis of classification efficiency throughout various taxa revealed that geNomad outperformed different instruments in all assessed teams (Supplementary Desk 5 and Supplementary Observe 3). Moreover, geNomad exhibited a decrease fee of misclassifying viruses as plasmids (1.7%) in comparison with all instruments besides PlasX (1.5–64.4%; Supplementary Desk 6 and Supplementary Observe 3).
In virus classification, geNomad attained the perfect general efficiency when contemplating all size strata (MCC: 95.3%, F1-score: 97.3%), adopted by VirSorter2 (ref. 30) executed with all fashions (MCC: 81.3%, F1-score: 88.9%), VirSorter2 executed with default parameters (MCC: 79.7%, F1-score: 87.1%) and PPR-Meta (MCC: 77.4%, F1-score: 86.6%) (Fig. 3b and Supplementary Desk 3). VIBRANT31, geNomad, VirSorter2 (default parameters) and DeepMicroClass achieved the best classification precision (97.5%, 97.3%, 94.7% and 92.6%, respectively), and Seeker32, DeepVirFinder33 and PPR-Meta obtained the bottom scores (61.8%, 80.5% and 88.5%, respectively).
In a benchmark research utilizing consultant genomes from the ICTV, we discovered that geNomad outperformed different instruments in all main taxa that we evaluated (Fig. 3c and Supplementary Desk 7). Notably, geNomad was the one device that achieved excessive sensitivity for viruses that encode an RNA-dependent RNA polymerase (RdRP; Orthornavirae, 98.64%) and large viruses (Megaviricetes, 94.74%) at a hard and fast FDR of 5%. When evaluating sensitivity throughout completely different host clades, we discovered that geNomad was the one device that recognized greater than 90% of the viruses infecting micro organism, archaea and a number of eukaryotic teams, whereas different instruments struggled to determine viruses that infect at the least two eukaryotic teams (Supplementary Desk 8). In a further benchmark the place we measured classification sensitivity on a catalog of metagenomic Inovirus34, that are recognized to be difficult to detect robotically, geNomad (sensitivity: 84.8%) additionally outperformed different evaluated instruments (common sensitivity: 32.5%) (Supplementary Desk 9).
We assessed the efficiency of geNomad’s taxonomic task (Fig. 3d and Supplementary Desk 10) by assigning 116,250 artificially fragmented genomes of ICTV exemplar species to viral lineages utilizing a marker dataset with modified taxonomic metadata to simulate novelty (see Strategies for particulars). Of the processed fragments, the bulk (80.3%) was efficiently assigned to a viral lineage, with most being categorized on the class (54.4%), order (13.6%) or household (10.1%) ranges. Amongst these, 48.2% have been accurately assigned to essentially the most particular rank (as much as the household stage); 49.5% have been under-classified (assigned to the right lineage however to not essentially the most particular rank); and solely 2.3% have been assigned to the mistaken lineage. These outcomes point out that geNomad is dependable at assigning sequences to larger taxa. The unassigned fragments, which lacked hits to markers with taxonomic info, have been principally shorter than 3 kb (80.6%).
Delicate and exact identification of proviruses
Temperate phages can combine into host genomes and kind proviruses, which may significantly have an effect on host metabolism and ecology35,36,37. To determine built-in viruses inside host genomes, geNomad employs a conditional random discipline (CRF) mannequin that identifies genomic areas that exhibit a excessive enrichment of viral markers and are flanked by chromosome markers (Fig. 4a). The CRF mannequin leverages the intensive gene protection supplied by the marker database and scores every gene, factoring within the specificity ranges of assigned markers for that gene and its neighboring genes. To remove spurious viral islands (areas of consecutive genes labeled as viral), geNomad merges carefully situated islands and subsequently removes these with a low marker enrichment—that’s, areas containing only some virus markers. Lastly, as a result of tRNAs and integrases are generally discovered subsequent to the sides of built-in components as a result of dynamics of site-specific recombination38, geNomad extends provirus boundaries up till neighboring tRNAs and/or integrases, bettering the detection sensitivity of genes near provirus edges.
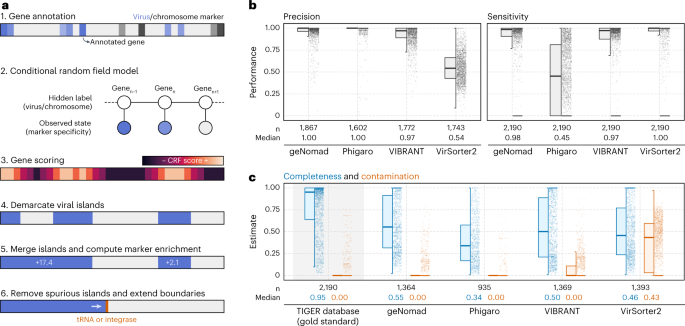
a, Provirus identification begins by annotating the genes inside a sequence with geNomad markers, which retailer info of how particular they’re to hosts or viruses. These specificity values are then fed to a CRF mannequin, which can rating every gene utilizing info from the markers in its environment. A rating cutoff is used to demarcate viral islands, and islands which can be shut collectively are merged. Islands with few viral markers are discarded, and the boundaries of the remaining islands are prolonged up till close by tRNAs or integrases. b, Distributions of the precision and sensitivity of a number of provirus identification instruments, measured on the gene stage for every provirus. Proviruses from the TIGER database have been used as the bottom reality for this benchmark. c, Completeness and contamination estimates of demarcated proviral areas that didn’t overlap with proviruses within the TIGER database. Estimates for TIGER proviruses are proven with a grey background as a reference. Field plots present the median (center line), interquartile vary (field boundaries) and 1.5 instances the interquartile vary (whiskers).
We evaluated geNomad’s provirus demarcation efficiency and in contrast it with different fashionable instruments (Phigaro39, VIBRANT and VirSorter2) utilizing the TIGER dataset38, which incorporates exactly mapped integration websites throughout 2,168 prokaryotic genomes, as the bottom reality (Fig. 4b and Supplementary Desk 11). For every predicted proviral area by the benchmarked instruments, we measured precision because the fraction of genes inside TIGER proviruses and sensitivity because the proportion of genes contained inside areas predicted by every device. The outcomes of this benchmark demonstrated that geNomad recognized extra proviruses than different instruments and exhibited excessive precision and sensitivity. Not all the expected proviral areas overlapped with TIGER coordinates, as a result of this dataset doesn’t embody inactive phages nor proviruses that don’t combine at tRNAs. To measure the standard of such predictions, we used CheckV40 (model 1.0.1) to estimate the standard of those areas and located that geNomad outperformed different instruments, because the proviruses it demarcated tended to be extra full with decrease contamination ranges (that’s, few host genes) (Fig. 4c and Supplementary Desk 11). The completeness of most of those proviral areas was comparatively decrease than these in TIGER, indicating that they doubtless characterize inactive proviruses that underwent gene loss. In a further benchmark, we discovered that geNomad outperforms different instruments within the identification of proviruses in a Pseudomonas aeruginosa pangenome41 (Supplementary Observe 4, Prolonged Knowledge Fig. 6 and Supplementary Desk 11).
geNomad is quick and permits evaluation of enormous datasets
To make geNomad accessible to a large viewers, we designed it to be user-friendly and environment friendly, permitting it to run shortly on a broad vary of {hardware}. geNomad could be put in domestically although various strategies (pip, Conda and Docker), facilitating its set up in a wide range of eventualities. The command line interface gives complete explanations and detailed execution logging. For non-technical customers, geNomad is offered as an online software by way of the NMDC EDGE platform ( permitting simple information add and outcome visualization within the internet browser. Moreover, the combination with NMDC EDGE permits geNomad to be simply integrated into bigger workflows that embody different duties, akin to meeting and binning.
In a benchmark measuring the time it took to categorise 10,000 metagenomic scaffolds, geNomad was quicker than all however two of the evaluated instruments (Desk 1), taking considerably much less time than VirSorter2 (26.1× enchancment), PlasX (8.1×), viralVerify (6.8×) and VIBRANT (2.7×). The one instruments that have been quicker have been DeepMicroClass and PlasClass, that are alignment-free instruments that exhibited decrease classification efficiency than geNomad in our benchmarks (Fig. 3a). It’s price noting that geNomad’s marker and sequence branches could be run independently, lowering runtime by half whereas nonetheless sustaining good classification efficiency (Supplementary Desk 3), in circumstances the place time is a priority. These outcomes reveal that, because of its pace, geNomad can be utilized in various {hardware} and could be scaled to course of giant datasets. Actually, geNomad was not too long ago used to course of roughly 260 million scaffolds (2.7 trillion base pairs) from IMG/M to assemble the info used to construct the IMG/VR model 4 (ref. 9) and IMG/PR databases, which characterize the biggest out there databases of virus and plasmid sequences, respectively.
geNomad permits the invention of RNA and large viruses
Latest research have unveiled a beforehand undiscovered variety of RNA viruses (Orthornavirae kingdom) and large viruses (Nucleocytoviricota phylum) by way of the evaluation of sequencing information from metatranscriptomes and metagenomes14,42,43,44,45,46. As current virus discovery instruments exhibit restricted efficacy in detecting a considerable fraction of the RNA and large virus genomes (Orthornavirae and Megaviricetes in Fig. 3c), these large-scale surveys have resorted to customized strategies, akin to figuring out the RdRP hallmark gene for RNA viruses and using metagenomic binning for large viruses. Nonetheless, these tailor-made approaches are sometimes tough to breed, as they have been developed for inside use. To handle this situation and improve the sensitivity of detecting each RNA and large viruses in sequencing information, we leveraged latest information about these viruses to coach geNomad, which improved the identification of those lineages (Fig. 3c, Supplementary Observe 5 and Supplementary Observe 6).
In metatranscriptomes from microbial communities of the Sand Creek Marshes47, geNomad categorized 99.9% of the sequences containing the RdRP gene as viral (Fig. 5a). Moreover, we discovered that 98.1% of the scaffolds that binned48 with RdRP-encoding sequences primarily based on their co-occurrence throughout a number of samples have been additionally recognized as viral by geNomad. This means that geNomad can determine RNA virus genome sequences even once they lack the RdRP gene (Fig. 5a). In distinction, different instruments categorized a median of solely 43.7% of those sequences as viral (Supplementary Desk 12). Inspection of pairs of co-occurring scaffolds revealed that they fell into two classes: (1) linear genomes that have been assembled into two scaffolds, one in all which lacked the RdRP gene (Marnaviridae bin in Fig. 5b); and (2) segmented genomes, containing a number of DNA molecules (Cystoviridae bin in Fig. 5b). Amongst sequences not encoding RdRP and never binned with RdRP-encoding scaffolds, but categorized as viruses by geNomad, we discovered fragments of RNA virus genomes lacking the RdRP gene (Leviviridae scaffold in Fig. 5b) and transcripts of DNA viruses (Caudoviricetes scaffold in Fig. 5b).
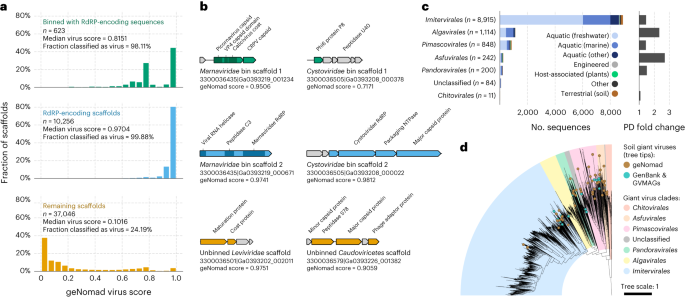
a, Histograms displaying the geNomad rating distribution of three teams of scaffolds of the Sand Creek Marshes metatranscriptomes: scaffolds that binned with RdRP-encoding sequences (high row, in inexperienced); scaffolds that include the RdRP gene (center row, in blue); and the remaining scaffolds (backside row, in orange). The median geNomad rating and the fraction of scaffolds categorized as viral are indicated for every group. b, Genome maps of chosen sequences that have been categorized as viral by geNomad. Two pairs of co-occurring Orthornavirae scaffolds are represented (Marnaviridae and Cystoviridae bins). Genes focused by geNomad markers are coloured, and genes that don’t match any marker are proven in grey. Rows and colours match these of a. c, Variety of scaffolds assigned to Nucleocytoviricota orders throughout a number of ecosystems (left bar plot). Sequences have been recognized by geNomad in a large-scale survey of metagenomes of various ecosystems. Solely scaffolds which can be at the least 50 kb lengthy or extra have been evaluated. Bar colours characterize the ecosystem varieties the place the sequences have been recognized. The phylogenetic variety (PD) fold change is proven on the best bar plot. PD fold change values correspond to the ratio between the entire PD of timber reconstructed with and with out geNomad-identified large viruses. d, Most probability phylogenetic tree of soil large viruses recognized with geNomad (brown tree ideas). Reference sequences from GenBank and from a earlier metagenomic survey (GVMAGs) have been included, and those that have been sequenced from soil samples are indicated with turquoise tree ideas. Tree ideas that aren’t coloured characterize consultant genomes sequenced from samples obtained from different ecosystems. The ranges equivalent to completely different Nucleocytoviricota orders are represented utilizing distinct colours.
To evaluate geNomad’s functionality to uncover new clades of large viruses, we utilized it to twenty-eight,865 metagenome assemblies from the IMG/M49 database. Scaffolds categorized as virus by geNomad that have been at the least 50 kb in size have been additional analyzed utilizing the GVClass pipeline, which positioned Nucleocytoviricota scaffolds in a phylogenetic context by figuring out a set of conserved protein households and reconstructing gene timber along with reference genomes. A complete of 11,414 scaffolds recognized by geNomad have been phylogenetically positioned within the Nucleocytoviricota tree (Fig. 5c and Supplementary Desk 13). Different instruments categorized, on common, 77.4% of those scaffolds as viral (Supplementary Desk 14). Inside metagenomes from soils, an understudied area of interest for large viruses50, we recognized 235 further Nucleocytoviricota scaffolds, up from 16 metagenomic bins reported within the earlier survey. Phylogenetic reconstruction of those soil large viruses revealed that they embody a number of novel clades of Imitervirales, Pimascovirales and Asfuvirales that do not need representatives in GenBank or Schulz et al.14 (Fig. 5d), suggesting that the underlying variety of Nucleocytoviricota in soil is significantly underestimated.
Extra info on the RNA and large virus surveys could be present in Supplementary Notes 5 and 6. The methodology is detailed in Supplementary Strategies.




